The use of genetic tests to diagnose and manage patients with myeloproliferative and myeloproliferative/myelodysplastic neoplasms, and related disorders
Methodology
This Good-Practice Paper was compiled according to the BSH process at https://b-s-h.org.uk/media/16732/bsh-guidance-development-process-dec-5-18.pdf. The British Society for Haematology (BSH) produces Good-Practice Papers to recommend good practice in areas where there is a limited evidence base but for which a degree of consensus or uniformity is likely to be beneficial to patient care. The Grading of Recommendations Assessment, Development and Evaluation (GRADE) nomenclature was used to evaluate levels of evidence and to assess the strength of recommendations. The GRADE criteria can be found at http://www.gradeworkinggroup.org.
Literature review details
Pubmed was searched from January 2018 to September 2020 using the terms (myeloproliferative OR polycythemia OR thrombocythemia OR myelofibrosis OR eosinophilia OR mastocytosis OR neutrophilia OR myelomonocytic OR eosinophilic CEL OR CNL or CMML or JMML) AND (mutation OR variant) AND (diagnosis OR prognosis). Summary information from the 1 063 hits was manually reviewed to identify 135 relevant publications. Relevant studies prior to January 2018 were identified from reviews published during the literature search period.
Review of the manuscript
Review of the manuscript was performed by the BSH Guidelines Committee General Haematology Task Force, the BSH Guidelines Committee and the General Haematology sounding board of BSH. It was also on the members section of the BSH website for comment. It has also been reviewed by members of the National Cancer Research Institute (NCRI) MPN subgroup, the Chair of the NCRI MDS subgroup and lead scientists from the Genomics Laboratory Hubs in England and representative genetic testing laboratories in Wales, Scotland and Northern Ireland; these organisations do not necessarily approve or endorse the contents.
Introduction
Genetics and genomics are playing an increasingly important role in the diagnosis and management of patients with haematological neoplasms. Next-generation sequencing (NGS) panels are widely available and initiatives such as the National Genomic Test Directory (NGTD; www.england.nhs.uk/publication/national-genomic-test-directories) in England along with parallel developments in the devolved nations aim to facilitate a standardised approach to testing and provide equity of access. A key component of this approach is the definition of eligibility criteria for specific tests to ensure appropriate usage from both clinical and financial perspectives.
This good-practice paper focuses on the use of genetic and genomic tests for adult chronic myeloid neoplasms as defined by the World Health Organization,1 including myeloproliferative neoplasms (MPN), myelodysplastic/myeloproliferative neoplasms (MDS/MPN), myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1–JAK2 (myeloid/lymphoid neoplasms with eosinophilia, MLN- eo) and mastocytosis. We have not included chronic myeloid leukaemia (CML) as this has been covered recently elsewhere,2 as has the full spectrum of clinical and laboratory investigations for patients with abnormal blood counts and/or suspected myeloid neoplasia.1, 3-9
Classical BCR–ABL1-negative myeloproliferative neoplasms
Screening investigations for erythrocytosis, thrombocytosis, suspected myelofibrosis and atypical thrombosis
Molecular screening investigations for the common MPN phenotype driver mutations (JAK2, CALR, MPL), usually performed on peripheral-blood DNA, are shown in Table I. These assays will identify a mutation in almost all patients with polycythaemia vera (PV) and 85–90% with essential thrombocythaemia (ET) and primary myelofibrosis (PMF). Single-target assays may be employed sequentially but multiplex assays, typically using NGS, sequence several targets in parallel and are more cost-effective. Either approach is acceptable if laboratory turnaround times and assay sensitivity10 are satisfactory [e.g. detection of 1–3% variant allele frequency (VAF) or lower for JAK2 c.1849G>T (p.Val617Phe), usually referred to as JAK2 V617F, and 5% VAF for JAK2 exon 12, CALR exon 9 or MPL exon 10 variants]. The use of broad myeloid NGS panels to screen cases with suspected MPN is unlikely to be cost-effective, but if larger panels are used we recommend that the initial analysis and report should be limited to common MPN driver mutations (Table I).
| Presentation | Variant | Frequency | References |
|---|---|---|---|
| Erythrocytosis | JAK2 V617F | 96–97% PV | 11, 12 |
| JAK2 exon 12 mutations* | ~3% PV | 12 | |
| Thrombocytosis | JAK2 V617F | 50–60% ET | 13, 14 |
| CALR exon 9 mutation | 25–30% ET | 15, 16 | |
| MPL exon 10 mutation | 3–11% ET | 17, 18 | |
| BCR–ABL1 fusion | To exclude CML | ||
| Suspected primary myelofibrosis | JAK2 V617F | 50–60% PMF | 16, 19 |
| CALR exon 9 mutation | 15–35% PMF | 15, 16 | |
| MPL exon 10 mutation | 6–9% PMF | 16, 18 | |
| Suspected chronic myeloid leukaemia | BCR–ABL1 fusion | 100% CML |
- ET, essential thrombocythaemia; PMF primary myelofibrosis; PV, polycythaemia vera.
- * Rare cases with a discrepancy between JAK2 exon 12 mutant allele burden in bone marrow and peripheral blood have been reported, so testing of bone marrow may be considered if there is a high index of suspicion.20
Universal reporting of mutant allele burden on diagnostic samples is not essential, although this should be considered where prognostically useful, e.g. suspected progression of PV to post-PV myelofibrosis (MF),21 or where demonstration of molecular response will be relevant (see subsection 1·3). Low allele burden results (e.g. <1% JAK2 V617F) should be reported as such, since the clinical significance may be less certain given the prevalence of low-level JAK2 V617F in the general population (see below). In patients with low-level JAK2 V617F and MPN phenotype, screening for CALR and MPL mutations should be carried out as these mutations may coexist.22 JAK2 V617F and CALR mutations may also coexist with BCR–ABL1, with such cases usually being identified following the persistence of thrombocytosis or other MPN features despite achievement of a good molecular response to tyrosine kinase inhibitor therapy for CML.23, 24 Specific CALR mutations (type 1, 52-bp deletion; type 2, 5-bp insertion; type 1-like and type 2-like)25 have prognostic significance in PMF (Table II) and should be reported routinely.
| Score | Disorder | Cytogenetic/molecular variable(s) included | HR | References |
|---|---|---|---|---|
| DIPSS+ | PMF | Unfavourable karyotype‡ | 2·4 | 26 |
| MYSEC-PM | Post-PV/post-ET MF | CALR-unmutated | 2·6 | 27 |
| MIPSS70 | PMF |
Absence of CALR type 1/type 1-like mutation At least 1 HMR* mutation 2 or more HMR* mutations |
1·89 1·77 3·95 |
28 |
| MIPSS70+ | PMF |
Absence of CALR type 1/type 1-like mutation At least 1 HMR* mutation 2 or more HMR* mutations Unfavourable karyotype§ |
2·4 1·8 2·4 3·1 |
28 |
| GIPSS | PMF |
Very high risk karyotype¶ Unfavourable karyotype¶ Absence of CALR type 1/type 1-like mutation ASXL1 mutation SRSF2 mutation U2AF1 mutation |
3·1 2·1 2·1 1·8 2·4 2·4 |
29 |
| MIPSS70+ v2 | PMF |
Very high risk karyotype¶ Unfavourable karyotype¶ 2 or more HMR mutations† 1 HMR mutation† Absence of type 1/type 1-like CALR mutation |
5·9 2·5 2·6 1·8 2·1 |
30 |
| Sanger multistage model | MPN |
Up to 53 genomic features (single gene variant/copy number information) |
31 | |
| MTSS | PMF/post-PV/post-ET MF |
Absence of CALR or MPL mutation ASXL1 mutation |
2·4 1·42 |
32 |
| FIM | PMF/post-PV/post-ET MF |
TP53 High risk mutations** ASXL1 only |
8·68 3·24 2·45 |
33 |
- ET, essential thrombocythaemia; MPN, myeloproliferative neoplasm; PMF primary myelofibrosis; PV, polycythaemia vera.
- * HMR, high molecular risk (ASXL1, IDH1/2, EZH2, SRSF2).
- † HMR, high molecular risk (ASXL1, IDH1/2, EZH2, SRSF2, U2AF1 Q157).
- ‡ Unfavourable: complex karyotype or sole or two abnormalities that include +8, −7/7q-, i(17q), −5/5q-, 12p-, inv(3) or 11q23 rearrangement.
- § Unfavourable: any abnormal karyotype other than sole abnormalities of 20q-, 13q-, +9, chromosome 1 translocation/duplication, −Y or sex chromosome abnormality other than −Y.
- ¶ Very high risk: single/multiple abnormalities of −7, i(17q), inv(3)/3q21, 12p-/12p11.2, 11q-/11q23, or other autosomal trisomies not including +8/ +9 (e.g. +21, +19); favourable: normal karyotype or sole abnormalities of 13q-, +9, 20q-, chromosome 1 translocation/duplication or sex chromosome abnormality including −Y; unfavourable: all other abnormalities.
- ** ≥1 mutation in EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS or KRAS. ASXL1-only mutations had no or limited prognostic value; however, ASXL1 mutations conferred a worse prognosis when associated with a mutation in TP53 or high-risk genes.
Clinical context must be considered prior to performing screening assays. In patients with erythrocytosis or thrombocytosis, molecular screening investigations (Table I) are recommended in those with persistently and significantly elevated counts (haematocrit >0·52 l/l in males or >0·48 l/l in females; platelet count ≥450 × 109/l),3, 4 after exclusion of secondary causes or where abnormalities are out of keeping with any possible secondary cause. Exclusion of BCR–ABL1 is important for all patients with thrombocytosis lacking a JAK2, CALR or MPL mutation or with atypical features (e.g. basophilia, left-shifted granulocytes, small hypolobated megakaryocytes). JAK2 V617F is also found in healthy individuals, at increasing prevalence with older age (‘clonal haematopoiesis’, CH).34-38 Although CH is associated with increased risk of developing cardiovascular disease,39 there is no prospective evidence to guide management of most patients with normal or near-normal blood counts who harbour JAK2 V617F but do not fulfil diagnostic criteria for MPN, even if there are also abnormalities on bone marrow histology. The JAK2 46/1 haplotype, and common polymorphisms in TERT and other genes only confer a weak predisposition to MPN and therefore there is no clinical value in screening for these in routine practice.40, 41
- Molecular screening for JAK2, CALR and MPL variants as appropriate (Table I) is recommended in patients with persistent erythrocytosis or thrombocytosis (GRADE 1B).
- Screening for JAK2 V617F is recommended in cases with normal blood counts and unexplained splanchnic vein thrombosis (GRADE 1B) and may be considered in selected patients with unexplained cerebral vein thrombosis (GRADE 2C).
- Screening for CALR variants may be considered in patients with splanchnic vein thrombosis or cerebral vein thrombosis (GRADE 2C).
- Screening for JAK2, CALR and MPL variants should be considered for patients with arterial or unprovoked venous thrombosis who have a mildly or variably elevated haematocrit or platelet count that persists for 2–3 months (GRADE 2C).
- BCR–ABL1 should be excluded in cases with persistent thrombocytosis negative for JAK2, CALR and MPL variants or with atypical features (GRADE 1B).
Testing for additional somatic driver variants with myeloid gene small-variant ‘panels’ +/− cytogenetic analysis
Additional somatic mutations in cancer driver genes include small variants (single nucleotide substitutions or small insertions/deletions) in TET2 (10–15% MPN), ASXL1 (5–10%) and DNMT3A (5–10%),15, 31, 48 all of which are also associated with CH.34-37 Mutations are found at lower prevalence in regulators of splicing (SRSF2, SF3B1, U2AF1, ZRSR2) and of chromatin structure, epigenetic functions and cellular signalling (e.g. EZH2, IDH1, IDH2, CBL, KRAS, NRAS, STAG2, TP53).31 Frequencies are often higher in PMF, post-PV or post-ET MF, and/or blast phase of other MPN or MDS/MPN.
The improved cost effectiveness of NGS technologies now permits widespread testing for panels of such ‘myeloid gene’ variants which, as a minimum for MPN, should include the genes listed under M85.2 in the NGTD (the current version can be found at https://www.england.nhs.uk/publication/national-genomic-test-directories/). There is a general consensus that reporting abnormalities down to 5% VAF is adequate for routine analysis, but standardised interpretation of panel results needs further development. For all myeloid neoplasms panel analysis can be performed with DNA extracted from peripheral blood, but DNA extracted from bone marrow is preferred if available. Running and reporting panels is relatively expensive, and in older populations can also identify incidental CH. Use of a panel for all MPN patients is therefore currently neither necessary nor easily deliverable, but panels can add useful supplementary information in specific situations, as detailed below.
Cytogenetic abnormalities are most often found in PMF or post-PV/post-ET MF, in which an abnormal karyotype is reported in up to 45% of patients.49, 50 Conventional karyotyping identifies the commoner copy number abnormalities and deletions (e.g. 20q-, 13q-, +8, +9, 1q+, −7/7q-) and less common balanced translocations [e.g. t(1;6)],51 and has been incorporated into several prognostic scoring systems.26, 29, 30 Other genome-wide technologies such as large pan-cancer NGS panels and SNP (single nucleotide polymorphism) arrays identify the common copy number losses and gains with greater resolution than conventional cytogenetics but will not identify balanced translocations. However these assays may also detect regions of copy-number-neutral loss of heterozygosity (LOH) that are not identified by conventional karyotyping but are included in some prognostic models.31 An abnormal karyotype is reported at diagnosis in 5–10% of patients with ET and ~15% with PV,52-54 and although such findings may have some prognostic significance, first line management is not generally altered as a result.
At presentation of a suspected MPN, with negative screening investigations
Erythrocytosis
Patients with unexplained erythrocytosis who lack JAK2 V617F may be considered for a bone marrow biopsy and JAK2 exon 12 mutation screening; most are diagnosed with ‘idiopathic’ erythrocytosis if there is no apparent secondary cause.3 The rare entity of JAK2-unmutated PV is still recognised in patients with other myeloproliferative clinicopathological features and marrow histology3 but its molecular aetiology is mostly undefined. A very small number of JAK2-unmutated cases with clonal erythrocytosis due to somatic mutations in the SH2B3 gene have been reported, although the phenotype was of idiopathic erythrocytosis with suppressed erythropoietin rather than classic PV55 and optimal management of such cases is unknown. There is currently insufficient evidence to recommend myeloid gene panel testing or cytogenetic analysis in the great majority of cases with JAK2-unmutated erythrocytosis. Testing may be considered in rare patients with true JAK2-unmutated PV, although there is no evidence to guide such practice. Other patients with JAK2-unmutated erythrocytosis may be considered for testing for congenital causes of erythrocytosis, as discussed elsewhere.3, 56
Thrombocytosis or suspected PMF
In the 10–15% of patients with ET and PMF who lack mutations in JAK2, CALR or MPL, the finding of an additional driver mutation in a myeloid gene panel can support the diagnosis of a clonal disorder, with the proviso that incidental CH could be found in older individuals. The likelihood of identifying a mutation in such patients depends on age, clinical presentation, and gene panel content. More than half of patients with ‘triple-negative’ PMF do harbour additional mutations when screened with comprehensive genomic assays31 and approximately a third have an abnormal karyotype.51 In patients with bone marrow histology and clinical features consistent with PMF, myeloid gene panel testing in combination with conventional karyotyping (or SNP array) is recommended.
The diagnosis of triple-negative ET is made on bone marrow histology, although distinction from reactive causes can be challenging, especially in those with mild thrombocytosis. A small minority harbour a non-canonical mutation in JAK2 or MPL, or in another driver gene.31, 57, 58 However, in a large analysis of recurrent genomic abnormalities in myeloid neoplasms, no mutations or chromosomal abnormalities were found in over 80% of patients with ‘triple-negative’ ET, including all those aged under 39 years.31 It remains possible that at least a subset of these patients may not have a clonal disorder.59 In older patients there is a higher likelihood of finding an additional driver mutation (or occasionally a chromosomal copy number abnormality or LOH, e.g. chromosome 20); however, the risk of incidental CH also increases. Other differential diagnoses including MDS/MPN should be considered in triple-negative patients with other ‘myeloid’ mutations, through correlation with blood counts and marrow appearances.
- Younger patients (e.g. under 60 years) with bone marrow histology typical of ET [or myeloproliferative neoplasm, unclassifiable (MPN-U) or suspected prefibrotic MF] where confirmation of a clonal disorder would be useful in view of the patient’s likely long-term disease course and ideally where a broad panel that covers non-canonical variants in JAK2 and MPL and a range of other driver genes is available.
- Patients with significant thrombocytosis (e.g. platelet count > 600 × 109/l), no reactive cause and borderline bone marrow histology, where cytoreduction would be indicated if there was convincing evidence of a clonal disorder. Examples would include those with an unexplained thrombotic event, particularly younger patients. For older patients without thrombosis, testing may be considered but results must be interpreted with caution in view of the possibility of incidental CH.
- A myeloid gene panel and cytogenetic analysis (or equivalent) is recommended for patients with bone marrow histology and clinical features consistent with PMF (+/− suggestive features of MDS or MDS/MPN) who test negative for JAK2/CALR/MPL (GRADE 1B).
- A myeloid gene panel and cytogenetic analysis (or equivalent) is not recommended for most patients with JAK2/CALR/MPL-negative erythrocytosis or thrombocytosis but may be considered in individual cases (GRADE 2C).
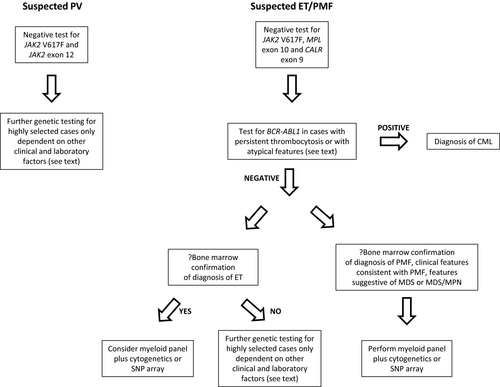
Patients with a known JAK2, CALR or MPL mutation
In patients with a confirmed clonal disorder, a myeloid gene panel and/or cytogenetic analysis can add information about diagnosis or prognosis at presentation or at suspected transformation; in future it may add information about options for targeted therapy.
Supplementary molecular information may allow definition of an alternative diagnosis associated with JAK2 V617F such as MDS/MPN. In patients with a clinical presentation suggestive of an MPN and a JAK2, CALR or MPL mutation, but with additional cytopenias(s) at diagnosis and unexplained ring sideroblasts or other morphological dysplasia, or with significant peripheral-blood monocytosis (monocytes ≥ 1 × 109/l), myeloid gene panel testing and cytogenetic analysis are recommended. The finding of other driver mutations may either support an alternative diagnosis (e.g. SF3B1 mutation in MDS/MPN with ring sideroblasts and thrombocytosis) or provide supportive information where the differential diagnosis is challenging [e.g. MPN with monocytosis versus chronic myelomonocytic leukaemia (CMML)].
For patients with ET and PV who develop cytopenias during cytoreductive therapy, marrow assessment may show morphological dysplasia with a differential diagnosis of disease progression versus therapy-related morphological changes. In this context myeloid gene panel testing and cytogenetic analysis may be considered. However, the finding of additional driver mutations is not evidence of disease progression per se in the absence of baseline molecular information. The number and nature of such variants must be considered in conjunction with a detailed clinical and drug history.
- Younger patients who at diagnosis fulfil BSH criteria for ET, PV or MPN-U but have atypical clinical features that warrant additional closer surveillance, e.g. marked splenomegaly, atypical bone marrow histology (including those meeting WHO criteria for prefibrotic MF).1
- Patients with MPN who are not candidates for allogeneic transplantation but in whom comprehensive prognostic information would aid clinical management and discussion with the patient.
- Patients requiring testing as part of entry to a clinical trial.
- Myeloid gene panel testing is recommended for MPN cases who test positive for JAK2/CALR/MPL mutations and have additional cytopenias(s) at diagnosis, unexplained ring sideroblasts or other dysplasia, increased blasts (including blastic transformation), peripheral-blood monocytosis or atypical clinical features (GRADE 1B).
- Myeloid gene panel testing and conventional karyotyping are recommended for all patients with PMF, post-PV or post-ET MF who are candidates for allogeneic stem cell transplant (GRADE 1B).
- Myeloid gene panel testing should be considered for other patients if the additional genomic data will guide clinical management (GRADE 2C).
Disease monitoring: quantitative assays of clonal burden
- High-sensitivity assays of mutant allele burden are recommended following post-allogeneic stem cell transplant to monitor for residual disease (GRADE 1C).
- Quantitative assays of mutant allele burden are not recommended for most MPN patients but may be considered where demonstration of molecular response would influence clinical management (GRADE 2C).
Atypical myeloproliferative neoplasms
CEL and MLN-eo
Patients with persistent eosinophilia of at least 1·5 × 109/l with no obvious secondary cause should be investigated for FIP1L1–PDGFRA on peripheral blood or bone marrow by FISH or nested reverse transcriptase (nested RT-) PCR.7, 78 Either technique alone may miss occasional cases79, 80 and so both, or other supplementary approaches,79, 81 should be considered in cases with a high index of suspicion.
Almost all tyrosine kinase (TK) gene fusions apart from FIP1L1–PDGFRA are associated with visible cytogenetic rearrangements and therefore bone marrow (BM) cytogenetic analysis should ideally be performed for cases with a suspected myeloid neoplasm if FIP1L1–PDGFRA is not detected.7, 78 The diversity of fusions precludes effective targeted RT-PCR analysis, although an increasing number of cases are being picked up by broad or targeted RNAseq screens. Although effective, this approach is currently too expensive to recommend as a general screening tool in all but exceptional cases. Break-apart FISH analysis for specific loci (PDGFRA, PDGFRB, FGFR1, JAK2 for MLN-eo; ABL1, FLT3, ETV6, other TK genes for CEL) may also be used to identify disruption of key loci. It is important that any suspected fusion (including FIP1L1–PDGFRA) identified by cytogenetics or FISH is confirmed by molecular methods to ensure that targeted therapy is used appropriately and to facilitate subsequent molecular monitoring, which is available for FIP1L1–PDGFRA and most other fusions in specialist centres. The timing of tests should follow that recommended for CML, including more frequent tests for patients attempting treatment-free remission.2, 82, 83
- Patients with persistent eosinophilia should be investigated initially for FIP1L1–PDGFRA by FISH and/or nested RT-PCR (GRADE 1B).
- BM cytogenetics or FISH is recommended to screen for other fusion genes, which must then be confirmed by molecular methods (GRADE 1B).
- Myeloid gene panel and KIT D816V testing should be considered for patients with persistent unexplained eosinophilia who test negative for fusion genes (GRADE 2B).
| Category | Genes | Frequency | References |
|---|---|---|---|
| MLN-eo | FIP1L1–PDGFRA | 5–20% HEUS; >80% MLN-eo | 84, 89 |
| Other PDGFRA fusions | Rare | ||
| PDGFRB fusions | <10% MLN-eo | ||
| FGFR1 fusions | <5% MLN-eo | ||
| PCM1–JAK2, BCR–JAK2 | <5% MLN-eo | ||
| Tyrosine kinase gene fusions in CEL and eosinophilia associated with other MPN or MDS/MPN | ETV6–ABL1 | ?1–2% HEUS/MPN-eo | |
| FLT3 fusions | Rare | ||
| Other JAK2 fusions | Rare | ||
| NTRK3, RET, ALK, others | Very rare | ||
| Other variants in CEL and eosinophilia associated with other MPN, MDS/MPN or SM | JAK2 V617F | 4% HEUS | 84 |
| JAK2 exon 13 indels | 1–2% HEUS | 88, 90 | |
| KIT D816V | 3% HEUS | 84 | |
| STAT5B N642H | 2% persistent eosinophilia including MPN-eo and MDS/MPN-eo | 87 | |
| DNMT3A, TET2, ASXL1, EZH2, SETBP1, CBL other myeloid genes | 11–21% HES/HEUS | 85, 86, 90 |
- HEUS, hypereosinophilia of undetermined significance; HES, idiopathic hypereosinophilic syndrome. MDS, myelodysplastic neoplasm; MPN, myeloproliferative neoplasm; SM, systemic mastocytosis.
CNL, MPN-U
CSF3R mutations are strongly, but not exclusively, associated with chronic neutrophilic leukaemia (CNL)91, 92 and are a central diagnostic feature of this disorder.1 Wider genomic profiling indicates a significant overlap in the pattern of mutated genes between CNL and MDS/MPN93 suggestive of a disease continuum. ASXL1 mutations were associated with an adverse prognosis in CNL in one study,94 but did not influence response to ruxolitinib.95
- Testing for CSF3R variants, preferably as part of wider myeloid panel, is recommended for all patients with suspected CNL (Grade 2B).
Mastocytosis
- Sensitive testing for KIT D816V is recommended for all patients with a clinical suspicion of mastocytosis (GRADE 1B).
- If negative for KIT D816V, screening for other KIT mutations should be considered for adults (but is recommended for children) (GRADE 1B).
Additional somatic mutations are found in 70–90% of advanced SM patients. Most mutation-positive cases have SM with an associated haematological neoplasm (SM-AHN), with the AHN usually being a subtype of MDS/MPN. Mutations are less frequent (<20%) in patients with indolent SM (ISM).101, 102 In advanced SM, mutations in SRSF2, ASXL1, RUNX1, EZH2 and NRAS have been associated with an adverse prognosis and thus molecular profiling is useful to guide transplant decisions.98, 102-104 In ISM, high VAF (≥30%) mutations in ASXL1, RUNX1 and/or DNMT3A have been associated with an adverse prognosis102 but the value of routine molecular profiling in this subtype remains to be established.
- Myeloid panel analysis is recommended for patients with advanced SM who are candidates for allogeneic stem cell transplantation (GRADE 1B).
- Myeloid panel analysis may be considered for other SM patients if the apparent aggressiveness of the disease might influence options for therapy (GRADE 2B).
- Myeloid panel and/or BM cytogenetics should be considered to characterise the AHN component of SM-AHN (GRADE 2B).
Myelodysplastic/myeloproliferative neoplasms
The diagnosis of the adult MDS/MPN overlap syndromes —CMML, atypical CML BCR–ABL1-negative (aCML), MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T) and MDS/MPN-unclassifiable (MDS/MPN-U) — remain heavily reliant on BM morphology and clinical assessment. Molecular genetics can, however, provide key information to assist with diagnosis, subclassification and prognostication across the spectrum of these disorders.
Initial investigations in suspected MDS/MPN
- BCR–ABL1 should be excluded in all cases of suspected MDS/MPN, and rearrangements associated with MLN-eo should be excluded in cases with eosinophilia (GRADE 1B).
Testing for additional somatic driver mutations with myeloid gene ‘panels’
In patients with indeterminate morphology
Somatic mutations are consistently reported to occur in >90% of cases across the MDS/MPN overlaps.112-115 The high frequency of somatic mutations in these conditions means the presence of a mutation can provide supportive evidence of clonality and assist in difficult diagnostic scenarios. Concerns have, however, been raised regarding the use of mutational analysis in this setting, due to reports of frequent somatic mutations in ageing healthy individuals.34-37 A recent study, however, in patients investigated for possible CMML, confirmed that even in the absence of definitive morphological features, those patients with a somatic mutation had a clinical phenotype and genotype indistinguishable from those with disease, and comparably poor outcomes.116 A myeloid gene panel is therefore recommended in difficult diagnostic cases and the presence of two mutations, one of which has a high VAF (>20%) would support a diagnosis.8 The genes included in the current NGTD for suspected MDS/MPN overlaps are listed in Table IV (see also NGTD test code M224.1) and the minimum genes recommended for the investigation of patients with suspected CMML in Table V. It is accepted that the genes included in the NGTD panel are a minimum requirement and larger panels may provide additional information, e.g. abnormalities of NPM1 are uncommon in MDS/MPN but identify cases likely to transform rapidly to AML, whereas abnormalities of FLT3 are potential therapeutic targets.8
| Pathway | Gene | aCML | CNL | CMML | MDS/MPN-RS-T | MDS/MPN-U |
|---|---|---|---|---|---|---|
| Signalling | KRAS | 3–10% | - | 7–23% | – | 4–5% |
| NRAS | 10–30% | 10% | 4–38% | – | 8–12% | |
| JAK2 | 4–11% | 8% | 1–11% | 37–78% | 8–25% | |
| CBL | 8–15% | 5% | 8–23% | 3% | 7–8% | |
| KIT | 6% | – | 0–3% | – | 4% | |
| FLT3 | 5–7% | - | 1–4% | – | 3–4% | |
| CSF3R | 0–25% | 60–80% | 2–4% | – | 4–6% | |
| SETBP1 | 7–38% | 14–56% | 4–18% | 1–6% | 13–16% | |
| SH2B3 | 0–4% | – | 0–5% | 3% | ||
| MPL | – | – | rare | 4–20% | 8% | |
| CALR | – | – | rare | 17% | 4% | |
| ETNK1 | 3–9% | 3% | 3–4% | 3% | 4% | |
| PTPN11 | 0–8% | ~0% | 3–5% | – | 4–5% | |
| NF1 | 0–4% | – | 6–10% | – | 4% | |
| Splicing | SF3B1 | 0–6% | 3% | 3–10% | 97% | 11–16% |
| SRSF2 | 37–48% | 44% | 24–55% | 4% | 24–48% | |
| U2AF1 | 3–15% | 15% | 2–24% | – | 8–19% | |
| ZRSR2 | 3–4% | 3% | 3–8% | – | 0–6% | |
| Transcription | RUNX1 | 6–20% | 3% | 8–28% | 1% | 4–17% |
| CEBPA | 4% | – | 0–20% | – | 4–8% | |
| GATA2 | 15–18% | 13% | 1–14% | 3% | 12–16% | |
| NPM1 | 4% | – | 1–3% | – | 0–3% | |
| BCOR | 4% | – | 3–7% | – | – | |
| CUX1 | 10–11% | 5% | 0–6% | 4% | 0–8% | |
| TP53 | 3% | 3% | 0–2% | 3% | 0–14% | |
| Cohesin | STAG2 | 11–15% | 3% | 3% | – | 8–16% |
| DNA methylation | DNMT3A | 4–7% | 5% | 2–12% | 18% | 0–13% |
| TET2 | 16–37% | 21% | 29–73% | 21% | 30–44% | |
| IDH1/2 | 0–3% | 3% | 1–7% | 3% | 0–10% | |
|
Histone modification |
ASXL1 | 28–92% | 57–77% | 32–69% | 0–11% | 53–64% |
| EZH2 | 13–33% | 21% | 5–13% | 7% | 10–25% |
- Data from [8, 92-94, 108, 115-119] and references therein. A dash indicates a mutation in that gene is rare or has not been reported. The NGTD also includes CHEK2, NFE2, IKZF1 and HRAS but the prevalence of mutations in these genes is unknown for CNL and MDS/MPN.
- MPN-U, myeloproliferative neoplasm, unclassifiable.
In patients with a confirmed diagnosis of MDS/MPN
The genes most commonly mutated in MDS/MPN are not specific for these conditions; however, genotypic/phenotypic correlations have been identified which can assist in subclassification. Mutations in genes with prognostic relevance can also be identified along with possible targets for therapy (JAK2, IDH1/2) with the latter likely to increase over time.
With respect to CMML, SRSF2, TET2 and ASXL1 are by far the most commonly mutated genes112-114 and the combination of mutation in TET2 and either SRSF2 or ZRSR2 is highly specific for a myelomonocytic phenotype.120 A diagnosis of aCML is supported by the presence of mutations in SETBP1 and/or ETNK1 which are reported in ~25–38% and ~10% of cases, respectively.117, 118, 121, 122 These genes are mutated less frequently in CMML and MDS/MPN-U although SETBP1 is also mutated in CNL.121, 122 Patients with aCML also show a relative lack of MPN phenotype driver mutations (JAK2, CALR, MPL)108, 123 with the presence of these tending to exclude this diagnosis.1 In MDS/MPN-RS-T, mutations in SF3B1 and JAK2 are reported in up to 90% and 57% of cases, with CALR or MPL mutations in a small minority119, 124, 125 and the detection of an SF3B1 mutation in patients with 15% ring sideroblasts can help define the diagnosis.1 Comutation of these genes would strongly support a diagnosis of MDS/MPN-RS-T though it is not a current requirement.1 Elevated tryptase and/or mast cell abnormalities in MDS/MPN suggests SM-AHN, which is often underdiagnosed but may be supported by the finding of KIT D816V.126 The detection of KIT D816V in the context of a confirmed MDS/MPN should trigger review of BM morphology for a possible co-existing mastocytosis.
Mutational analysis is now incorporated into prognostic scoring systems across these diseases. Four genes (ASXL1, NRAS, RUNX1 and SETBP1) are independently associated with a worse overall survival (OS) in CMML and have been incorporated into the most recent CMML-specific prognostic scoring system (CPSS)-molecular and analysis of these is defined as mandatory for risk assessment.8 The number of mutations per patient has also been shown to correlate inversely with OS,112 and ASXL1 and/or NRAS mutations are associated with worse survival after stem cell transplantation.127 ASXL1 and SETBP1 also infer a poor prognosis across other MDS/MPN with these genes being commonly comutated.115, 121, 122, 128 In atypical CML, SETBP1 was associated with an adverse clinical phenotype and a significantly worse OS121, 122 while both SETBP1 and ASXL1 were associated with poor survival in patients with MDS/MPN-RS-T and have been incorporated into a mutation-enhanced prognostic model.128
- Myeloid gene panel analysis and BM cytogenetics or SNP array is recommended for patients diagnosed with MDS/MPN and for cases with suspected MDS/MPN but with indeterminate morphology (GRADE 1B).
Future directions
The landscape of genetic and genomic testing is changing rapidly, with broad screening techniques such as large pan-cancer panels, whole-genome sequencing and RNAseq beginning to impact on routine practice. Genomic, transcriptomic and epigenetic profiling of single cells are providing novel insights into the complexity and diversity of clonal disorders. Whilst these approaches clearly have huge potential, e.g. by facilitating comprehensive prognostic modelling,62 detection of rare targetable gene fusions129 and potentially cell type-specific assessment of measurable residual disease,130 it is currently unclear when or whether they will be cost-effective compared to more diverse, targeted approaches.
Acknowledgements
The BSH General Haematology task force members at the time of writing this good-practice paper were Mamta Garg (Chair), Charlotte Bradbury, Barbara De La Salle, Emmy Dickens, Savio Fernandes, John Grainger. Ciaran Mooney, Noemi Roy, Sara Stuart-Smith and Nicola Svenson. The authors would like to thank them, the BSH sounding board and the BSH guidelines committee for their support in preparing this good-practice paper.
Conflicts of interest
All authors have made a declaration of interests to the BSH and Task Force Chairs which may be viewed on request. NCPC has received honoraria for advisory boards and research support from Novartis and advisory boards from Incyte; ALG and CC have received honoraria as speakers for Novartis and advisory boards for AOP Orphan Pharmaceuticals; AJM has received honoraria from Novartis, Bristol Myers Squibb/Celgene, CTI BioPharma and AbbVie and has received research funding from Novartis, Bristol Myers Squibb/Celgene, and CTI BioPharma. MG has no conflicts of interest to declare.
Review process
Members of the writing group will inform the writing group Chair if any new pertinent evidence becomes available that would alter the strength of the recommendations made in this document or render it obsolete. The document will be archived and removed from the BSH current guidelines website if it becomes obsolete. If new recommendations are made an addendum will be published on the BSH guidelines website (www.b-s-h.org.uk).
Disclaimer
While the advice and information in this guidance is believed to be true and accurate at the time of going to press, neither the authors, the BSH nor the publishers accept any legal responsibility for the content of this guidance.